Code
librarian::shelf(
dplyr,
glue,
here,
tidyr
)In 2026 management of this data is being transferred to SEACAR In this file we prepare a transformation of the dataset for SEACAR ingestion, and perform some checks.
The data here should align with standards described in SEACAR_Metadata_UnifiedWQ_ID10006.xlsx.
library(dplyr)
library(scales)
# show percentage of missing or na rows
# 1. Get the total number of rows for each program
total_counts <- df %>%
count(ProgramName, name = "total_rows")
# 2. Get the number of NA rows for each program
na_counts <- df %>%
filter(is.na(ProgramID)) %>%
count(ProgramName, name = "missing_rows")
# 3. Combine total and NA counts, calculate percentage
percentage_table <- total_counts %>%
left_join(na_counts, by = "ProgramName") %>%
mutate(
# If a program had no NAs, na_counts will be NA. Replace with 0.
missing_rows = ifelse(is.na(missing_rows), 0, missing_rows),
# Calculate the percentage
percentage_raw = missing_rows / total_rows,
# Format the percentage for clean table output
percentage = scales::percent(percentage_raw, accuracy = 0.1)
) %>%
# Select and order the columns for the final table
select(ProgramName, missing_rows, total_rows, percentage) %>%
# Sort by program name
arrange(ProgramName)
# 4. Print the final data frame (which R displays as a table)
print(percentage_table) ProgramName missing_rows total_rows percentage
1 AOML_FBBB 0 43401 0.0%
2 BBAP 0 10594 0.0%
3 BBWW 0 910 0.0%
4 BROWARD 0 15142 0.0%
5 BROWARD_STORET 0 24290 0.0%
6 DEP 0 99792 0.0%
7 DERM_BBWQ 0 144946 0.0%
8 DERM_BBWQ_STORET 0 442825 0.0%
9 FIU_Estuaries 0 388128 0.0%
10 FIU_WQMP 0 62621 0.0%
11 FIU_WQMP_HISTORICAL 0 484218 0.0%
12 FIU_WQMP_RECENT 0 4020 0.0%
13 MiamiBeach 0 27484 0.0%
14 PALMBEACH 0 7658 0.0%
15 PALMBEACH_STORET 0 13145 0.0%
16 SFER 0 76230 0.0%library(dplyr)
library(scales)
# show percentage of missing or na rows
# 1. Get the total number of rows for each program
total_counts <- df %>%
count(ProgramName, name = "total_rows")
# 2. Get the number of NA rows for each program
na_counts <- df %>%
filter(is.na(SampleDate)) %>%
count(ProgramName, name = "missing_rows")
# 3. Combine total and NA counts, calculate percentage
percentage_table <- total_counts %>%
left_join(na_counts, by = "ProgramName") %>%
mutate(
# If a program had no NAs, na_counts will be NA. Replace with 0.
missing_rows = ifelse(is.na(missing_rows), 0, missing_rows),
# Calculate the percentage
percentage_raw = missing_rows / total_rows,
# Format the percentage for clean table output
percentage = scales::percent(percentage_raw, accuracy = 0.1)
) %>%
# Select and order the columns for the final table
select(ProgramName, missing_rows, total_rows, percentage) %>%
# Sort by program name
arrange(ProgramName)
# 4. Print the final data frame (which R displays as a table)
print(percentage_table) ProgramName missing_rows total_rows percentage
1 AOML_FBBB 0 43401 0.0%
2 BBAP 0 10594 0.0%
3 BBWW 0 910 0.0%
4 BROWARD 0 15142 0.0%
5 BROWARD_STORET 0 24290 0.0%
6 DEP 0 99792 0.0%
7 DERM_BBWQ 0 144946 0.0%
8 DERM_BBWQ_STORET 0 442825 0.0%
9 FIU_Estuaries 0 388128 0.0%
10 FIU_WQMP 0 62621 0.0%
11 FIU_WQMP_HISTORICAL 0 484218 0.0%
12 FIU_WQMP_RECENT 0 4020 0.0%
13 MiamiBeach 0 27484 0.0%
14 PALMBEACH 0 7658 0.0%
15 PALMBEACH_STORET 0 13145 0.0%
16 SFER 18 76230 0.0%library(dplyr)
library(scales)
# show percentage of missing or na rows
# 1. Get the total number of rows for each program
total_counts <- df %>%
count(ProgramName, name = "total_rows")
# 2. Get the number of NA rows for each program
na_counts <- df %>%
filter(is.na(ResultValue)) %>%
count(ProgramName, name = "missing_rows")
# 3. Combine total and NA counts, calculate percentage
percentage_table <- total_counts %>%
left_join(na_counts, by = "ProgramName") %>%
mutate(
# If a program had no NAs, na_counts will be NA. Replace with 0.
missing_rows = ifelse(is.na(missing_rows), 0, missing_rows),
# Calculate the percentage
percentage_raw = missing_rows / total_rows,
# Format the percentage for clean table output
percentage = scales::percent(percentage_raw, accuracy = 0.1)
) %>%
# Select and order the columns for the final table
select(ProgramName, missing_rows, total_rows, percentage) %>%
# Sort by program name
arrange(ProgramName)
# 4. Print the final data frame (which R displays as a table)
print(percentage_table) ProgramName missing_rows total_rows percentage
1 AOML_FBBB 0 43401 0.0%
2 BBAP 3 10594 0.0%
3 BBWW 0 910 0.0%
4 BROWARD 0 15142 0.0%
5 BROWARD_STORET 1410 24290 5.8%
6 DEP 0 99792 0.0%
7 DERM_BBWQ 15 144946 0.0%
8 DERM_BBWQ_STORET 2850 442825 0.6%
9 FIU_Estuaries 21510 388128 5.5%
10 FIU_WQMP 2773 62621 4.4%
11 FIU_WQMP_HISTORICAL 121735 484218 25.1%
12 FIU_WQMP_RECENT 773 4020 19.2%
13 MiamiBeach 347 27484 1.3%
14 PALMBEACH 44 7658 0.6%
15 PALMBEACH_STORET 329 13145 2.5%
16 SFER 14482 76230 19.0%library(dplyr)
library(scales)
# show percentage of missing or na rows in df$Monitoring.Location.ID grouped by df$program
# 1. Get the total number of rows for each program
total_counts <- df %>%
count(ProgramName, name = "total_rows")
# 2. Get the number of NA rows for each program
na_counts <- df %>%
filter(is.na(OriginalLatitude) | is.na(OriginalLongitude)) %>%
count(ProgramName, name = "missing_rows")
# 3. Combine total and NA counts, calculate percentage
percentage_table <- total_counts %>%
left_join(na_counts, by = "ProgramName") %>%
mutate(
# If a program had no NAs, na_counts will be NA. Replace with 0.
missing_rows = ifelse(is.na(missing_rows), 0, missing_rows),
# Calculate the percentage
percentage_raw = missing_rows / total_rows,
# Format the percentage for clean table output
percentage = scales::percent(percentage_raw, accuracy = 0.1)
) %>%
# Select and order the columns for the final table
select(ProgramName, missing_rows, total_rows, percentage) %>%
# Sort by program name
arrange(ProgramName)
# 4. Print the final data frame (which R displays as a table)
print(percentage_table) ProgramName missing_rows total_rows percentage
1 AOML_FBBB 0 43401 0.0%
2 BBAP 0 10594 0.0%
3 BBWW 0 910 0.0%
4 BROWARD 0 15142 0.0%
5 BROWARD_STORET 24290 24290 100.0%
6 DEP 261 99792 0.3%
7 DERM_BBWQ 0 144946 0.0%
8 DERM_BBWQ_STORET 442825 442825 100.0%
9 FIU_Estuaries 0 388128 0.0%
10 FIU_WQMP 0 62621 0.0%
11 FIU_WQMP_HISTORICAL 54 484218 0.0%
12 FIU_WQMP_RECENT 4020 4020 100.0%
13 MiamiBeach 4443 27484 16.2%
14 PALMBEACH 7658 7658 100.0%
15 PALMBEACH_STORET 13145 13145 100.0%
16 SFER 18 76230 0.0%library(dplyr)
library(scales)
# show percentage of missing or na rows
# 1. Get the total number of rows for each program
total_counts <- df %>%
count(ProgramName, name = "total_rows")
# 2. Get the number of NA rows for each program
na_counts <- df %>%
filter(is.na(ProgramLocationID)) %>%
count(ProgramName, name = "missing_rows")
# 3. Combine total and NA counts, calculate percentage
percentage_table <- total_counts %>%
left_join(na_counts, by = "ProgramName") %>%
mutate(
# If a program had no NAs, na_counts will be NA. Replace with 0.
missing_rows = ifelse(is.na(missing_rows), 0, missing_rows),
# Calculate the percentage
percentage_raw = missing_rows / total_rows,
# Format the percentage for clean table output
percentage = scales::percent(percentage_raw, accuracy = 0.1)
) %>%
# Select and order the columns for the final table
select(ProgramName, missing_rows, total_rows, percentage) %>%
# Sort by program name
arrange(ProgramName)
# 4. Print the final data frame (which R displays as a table)
print(percentage_table) ProgramName missing_rows total_rows percentage
1 AOML_FBBB 0 43401 0.0%
2 BBAP 0 10594 0.0%
3 BBWW 0 910 0.0%
4 BROWARD 0 15142 0.0%
5 BROWARD_STORET 0 24290 0.0%
6 DEP 0 99792 0.0%
7 DERM_BBWQ 0 144946 0.0%
8 DERM_BBWQ_STORET 0 442825 0.0%
9 FIU_Estuaries 0 388128 0.0%
10 FIU_WQMP 0 62621 0.0%
11 FIU_WQMP_HISTORICAL 0 484218 0.0%
12 FIU_WQMP_RECENT 0 4020 0.0%
13 MiamiBeach 0 27484 0.0%
14 PALMBEACH 0 7658 0.0%
15 PALMBEACH_STORET 0 13145 0.0%
16 SFER 18 76230 0.0%library(dplyr)
library(scales)
# show percentage of missing or na rows
# 1. Get the total number of rows for each program
total_counts <- df %>%
count(ProgramName, name = "total_rows")
# 2. Get the number of NA rows for each program
na_counts <- df %>%
filter(is.na(ParameterUnits)) %>%
count(ProgramName, name = "missing_rows")
# 3. Combine total and NA counts, calculate percentage
percentage_table <- total_counts %>%
left_join(na_counts, by = "ProgramName") %>%
mutate(
# If a program had no NAs, na_counts will be NA. Replace with 0.
missing_rows = ifelse(is.na(missing_rows), 0, missing_rows),
# Calculate the percentage
percentage_raw = missing_rows / total_rows,
# Format the percentage for clean table output
percentage = scales::percent(percentage_raw, accuracy = 0.1)
) %>%
# Select and order the columns for the final table
select(ProgramName, missing_rows, total_rows, percentage) %>%
# Sort by program name
arrange(ProgramName)
# 4. Print the final data frame (which R displays as a table)
print(percentage_table) ProgramName missing_rows total_rows percentage
1 AOML_FBBB 0 43401 0.0%
2 BBAP 0 10594 0.0%
3 BBWW 0 910 0.0%
4 BROWARD 0 15142 0.0%
5 BROWARD_STORET 0 24290 0.0%
6 DEP 0 99792 0.0%
7 DERM_BBWQ 0 144946 0.0%
8 DERM_BBWQ_STORET 0 442825 0.0%
9 FIU_Estuaries 0 388128 0.0%
10 FIU_WQMP 0 62621 0.0%
11 FIU_WQMP_HISTORICAL 0 484218 0.0%
12 FIU_WQMP_RECENT 4020 4020 100.0%
13 MiamiBeach 0 27484 0.0%
14 PALMBEACH 0 7658 0.0%
15 PALMBEACH_STORET 0 13145 0.0%
16 SFER 76230 76230 100.0%#show table from df with a row for each unique of
# (ProgramLocationID and ProgramName) with columns
# programLocationID, ProgramName, and N_locations,
# where N_locations is the number of unique OriginalLatitude and
# OriginalLongitude pairs for the programLocationID+ProgramName.
library(dplyr)
summary_table <- df %>%
group_by(ProgramLocationID, ProgramName) %>%
summarise(
N_locations = n_distinct(paste(OriginalLatitude, OriginalLongitude)),
.groups = "drop"
) %>%
filter(N_locations > 1)
summary_table# A tibble: 292 × 3
ProgramLocationID ProgramName N_locations
<chr> <chr> <int>
1 1 AOML_FBBB 133
2 1 SFER 51
3 10 AOML_FBBB 113
4 10 SFER 53
5 11 AOML_FBBB 117
6 11 SFER 55
7 12 AOML_FBBB 116
8 12 SFER 57
9 13 AOML_FBBB 119
10 13 SFER 54
# ℹ 282 more rows| Name | df |
| Number of rows | 1845404 |
| Number of columns | 35 |
| _______________________ | |
| Column type frequency: | |
| character | 18 |
| logical | 1 |
| numeric | 16 |
| ________________________ | |
| Group variables | None |
Variable type: character
| skim_variable | n_missing | complete_rate | min | max | empty | n_unique | whitespace |
|---|---|---|---|---|---|---|---|
| Habitat | 1480700 | 0.20 | 12 | 21 | 0 | 3 | 0 |
| IndicatorName | 1821453 | 0.01 | 9 | 13 | 0 | 3 | 0 |
| ManagedAreaName | 1821453 | 0.01 | 29 | 38 | 0 | 2 | 0 |
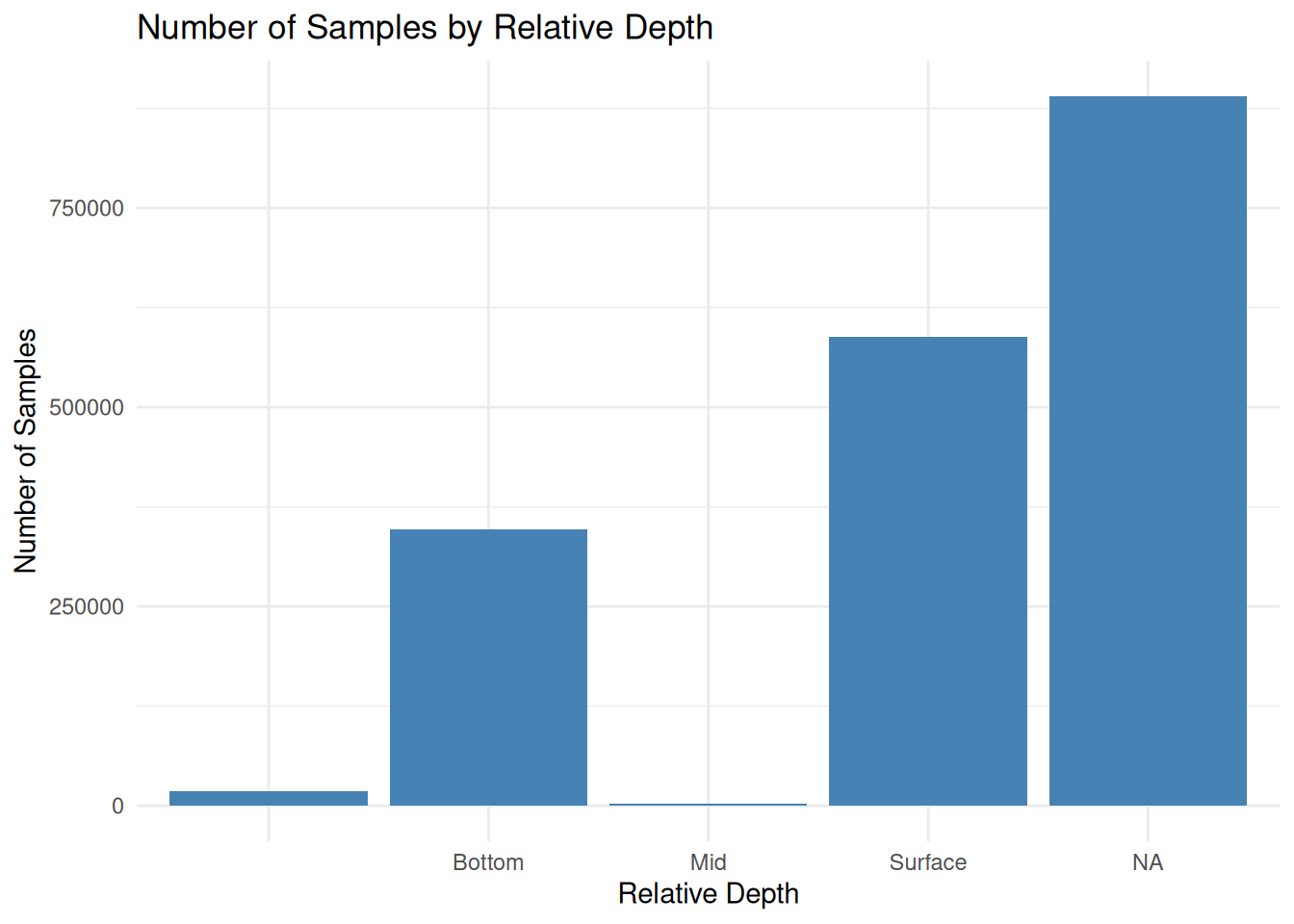
| RelativeDepth | 890081 | 0.52 | 0 | 7 | 17934 | 4 | 0 |
| DetectionUnit | 1843238 | 0.00 | 4 | 4 | 0 | 1 | 0 |
| SEACAR_QAQCFlagCode | 1821453 | 0.01 | 2 | 13 | 0 | 26 | 0 |
| SEACAR_QAQC_Description | 1821453 | 0.01 | 31 | 195 | 0 | 26 | 0 |
| ExportVersion | 1821453 | 0.01 | 23 | 23 | 0 | 3 | 0 |
| Region | 1778072 | 0.04 | 2 | 19 | 0 | 22 | 0 |
| ProgramName | 0 | 1.00 | 3 | 19 | 0 | 16 | 0 |
| ParameterName | 0 | 1.00 | 2 | 41 | 0 | 20 | 0 |
| ParameterUnits | 80250 | 0.96 | 0 | 10 | 14862 | 20 | 0 |
| ProgramLocationID | 18 | 1.00 | 1 | 25 | 0 | 2133 | 0 |
| ActivityType | 1503741 | 0.19 | 5 | 15 | 0 | 4 | 0 |
| SampleDate | 18 | 1.00 | 10 | 10 | 0 | 3702 | 0 |
| ValueQualifier | 1305542 | 0.29 | 0 | 4 | 221265 | 95 | 0 |
| ResultComments | 1504579 | 0.18 | 0 | 874 | 308298 | 756 | 9 |
| SEACAR_EventID | 1822363 | 0.01 | 36 | 36 | 0 | 2394 | 0 |
Variable type: logical
| skim_variable | n_missing | complete_rate | mean | count |
|---|---|---|---|---|
| ValueQualifierSource | 1845404 | 0 | NaN | : |
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| RowID | 1822363 | 0.01 | 1009723.34 | 798651.12 | 486217.00 | 514368.00 | 534968.00 | 1551534.00 | 3870318.00 | ▇▂▁▁▁ |
| ProgramID | 0 | 1.00 | 2165.93 | 2360.54 | 3.00 | 297.00 | 509.00 | 4018.00 | 10013.00 | ▇▁▆▁▁ |
| IndicatorID | 1821453 | 0.01 | 6.73 | 0.63 | 6.00 | 6.00 | 7.00 | 7.00 | 8.00 | ▆▁▇▁▂ |
| ParameterID | 1821453 | 0.01 | 8.60 | 6.56 | 1.00 | 3.00 | 6.00 | 16.00 | 19.00 | ▇▃▁▂▅ |
| AreaID | 1821453 | 0.01 | 6.09 | 0.89 | 6.00 | 6.00 | 6.00 | 6.00 | 15.00 | ▇▁▁▁▁ |
| Year | 972959 | 0.47 | 2010.98 | 9.77 | 1989.48 | 2002.08 | 2016.00 | 2020.00 | 2024.00 | ▁▃▃▁▇ |
| Month | 972941 | 0.47 | 6.59 | 3.44 | 0.00 | 4.00 | 7.00 | 10.00 | 12.00 | ▅▅▇▆▇ |
| TotalDepth_m | 1845341 | 0.00 | 62.22 | 105.84 | 1.10 | 2.10 | 2.85 | 8.50 | 250.00 | ▇▁▁▁▂ |
| MDL | 1657062 | 0.10 | 0.06 | 0.49 | 0.00 | 0.00 | 0.01 | 0.08 | 50.50 | ▇▁▁▁▁ |
| PQL | 1657062 | 0.10 | 0.18 | 2.41 | 0.00 | 0.01 | 0.06 | 0.20 | 250.00 | ▇▁▁▁▁ |
| Include | 1821453 | 0.01 | 1.00 | 0.03 | 0.00 | 1.00 | 1.00 | 1.00 | 1.00 | ▁▁▁▁▇ |
| MADup | 1821453 | 0.01 | 1.00 | 0.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | ▁▁▇▁▁ |
| ResultValue | 166271 | 0.91 | 11.01 | 389.59 | -0.03 | 0.03 | 0.61 | 6.00 | 53465.00 | ▇▁▁▁▁ |
| ActivityDepth_m | 991216 | 0.46 | 6.28 | 14.51 | 0.00 | 0.50 | 3.20 | 8.49 | 2494.00 | ▇▁▁▁▁ |
| OriginalLatitude | 496660 | 0.73 | 25.35 | 0.62 | 24.38 | 24.83 | 25.28 | 25.78 | 28.78 | ▇▇▁▁▁ |
| OriginalLongitude | 496714 | 0.73 | -80.98 | 0.74 | -85.02 | -81.52 | -80.93 | -80.29 | -80.02 | ▁▁▁▆▇ |
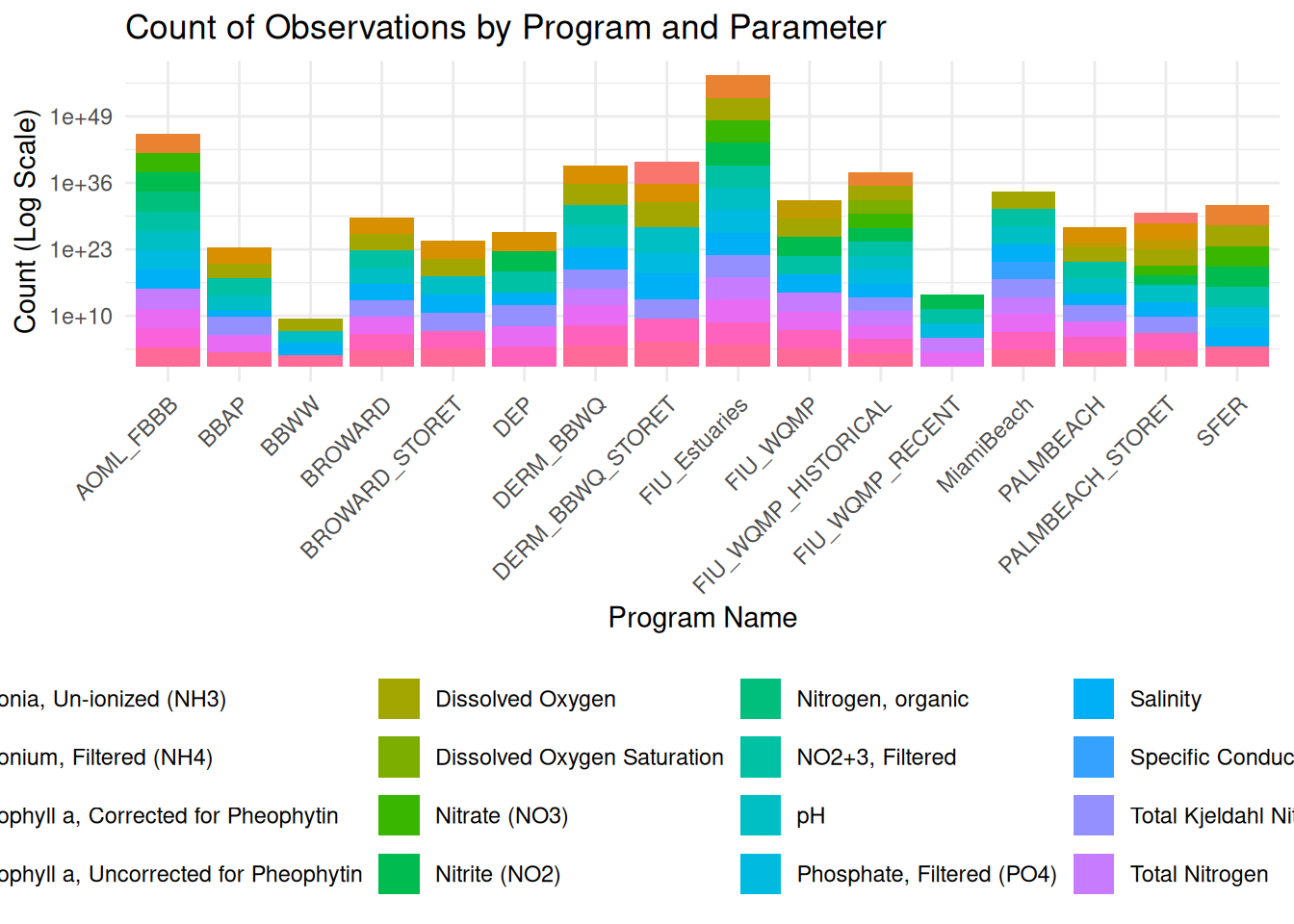
library(ggplot2)
ggplot(df, aes(x = ProgramName, fill = ParameterName)) +
geom_bar() +
scale_y_log10() +
labs(
title = "Count of Observations by Program and Parameter",
x = "Program Name",
y = "Count (Log Scale)",
fill = "Parameter"
) +
theme_minimal() +
theme(
legend.position = "bottom",
axis.text.x = element_text(angle = 45, hjust = 1)
)
source(here("R/mutateWINToSEACAR.R"))
df <- df_all %>% mutateWINToSEACAR(.keep = "unused")
SEACAR_COLUMNS <- c(
"ExportVersion","RowID","ProgramID","ProgramName","Habitat",
"IndicatorID","IndicatorName","ParameterID","ParameterName",
"ParameterUnits","ProgramLocationID","AreaID","ManagedAreaName",
"Region","ActivityType","SampleDate","ResultValue","Year","Month",
"ActivityDepth_m","RelativeDepth","TotalDepth_m","MDL","PQL",
"DetectionUnit","ValueQualifier","ValueQualifierSource",
"SampleFraction","ResultComments","OriginalLatitude",
"OriginalLongitude","SEACAR_QAQCFlagCode",
"SEACAR_QAQC_Description","Include","SEACAR_EventID","MADup"
)
# figure out which are actually present
present_cols <- intersect(SEACAR_COLUMNS, names(df))
missing_cols <- setdiff(SEACAR_COLUMNS, names(df))
if (length(missing_cols) > 0) {
warning("\nThese SEACAR_COLUMNS were not found in df:\n ",
paste(missing_cols, collapse = ", "))
}
# Any additional columns in df that are not mapped to SEACAR
extra_columns <- setdiff(names(df), SEACAR_COLUMNS)
cat("\nColumns from source data that are not (directly) mapped to SEACAR columns:\n")
Columns from source data that are not (directly) mapped to SEACAR columns:character(0)