load libraries
librarian::shelf(
dplyr,
tidyr,
here,
DT
)The allData.csv file is in “long” format, with “analyte name” as a column. This document transforms the data to “wide” “tidy” format, with “analyte name” on the columns. This is done by pivoting on the analyte name column.
Long format dimensions: Rows: 1265123 Columns: 10 Before pivoting we assess how many collisions will occur.
# A tibble: 249,180 × 6
source site datetime sample_depth analyte n
<chr> <chr> <chr> <dbl> <chr> <int>
1 AOML_FBBB 1 1998-01-20 NA Salinity 2
2 AOML_FBBB 1 1998-01-20 NA Water Temperature 2
3 AOML_FBBB 1 1998-03-10 NA Salinity 2
4 AOML_FBBB 1 1998-03-10 NA Water Temperature 2
5 AOML_FBBB 1 1998-06-30 NA Salinity 2
6 AOML_FBBB 1 1998-06-30 NA Water Temperature 2
7 AOML_FBBB 14 1998-01-21 NA Salinity 2
8 AOML_FBBB 14 1998-01-21 NA Water Temperature 2
9 AOML_FBBB 16 1998-12-21 NA Ammonium, Filtered (NH4) 2
10 AOML_FBBB 16 1998-12-21 NA NO2+3, Filtered 2
# ℹ 249,170 more rowsThese rows will collide with n other rows (see column on far right). An example cause of this is samples that were taken at the same time on the same day. Collisions in this pivot are averaged with a mean.
The transformation uses tidyr::pivot_wider() to convert the dataset from long format (where each row is one measurement of one analyte) to wide format (where each row represents all measurements at a specific location and time).
library(dplyr)
library(tidyr)
# Pivot to wide format using the actual column names from allData.csv
# Columns: source, site, datetime, analyte, value, units, latitude, longitude, sample_depth
df_wide <- df_long %>%
pivot_wider(
id_cols = c(source, site, datetime, units, latitude, longitude, sample_depth),
names_from = analyte,
values_from = value,
values_fn = mean # aggregate duplicates by taking mean
)
# Show dimensions of wide format data
cat("Wide format dimensions:\n")Wide format dimensions: Rows: 354239 Columns: 27
Number of analyte columns created: 20 Summary of numeric analyte columns:| Name | df_wide[, numeric_cols] |
| Number of rows | 354239 |
| Number of columns | 23 |
| _______________________ | |
| Column type frequency: | |
| numeric | 23 |
| ________________________ | |
| Group variables | None |
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| latitude | 157882 | 0.55 | 25.63 | 0.51 | 24.40 | 25.29 | 25.74 | 25.88 | 28.78 | ▃▇▁▁▁ |
| longitude | 157882 | 0.55 | -80.49 | 0.52 | -85.02 | -80.62 | -80.28 | -80.16 | -80.02 | ▁▁▁▁▇ |
| sample_depth | 198581 | 0.44 | 0.44 | 0.20 | 0.00 | 0.50 | 0.50 | 0.50 | 1.00 | ▃▁▇▂▁ |
| Water Temperature | 263170 | 0.26 | 19.55 | 11.63 | 0.00 | 2.10 | 24.60 | 28.45 | 74.65 | ▃▇▂▁▁ |
| Salinity | 266376 | 0.25 | 18.52 | 16.10 | 0.00 | 1.16 | 22.72 | 34.57 | 933.00 | ▇▁▁▁▁ |
| Dissolved Oxygen | 276439 | 0.22 | 4.45 | 11.33 | 0.00 | 1.51 | 5.05 | 6.45 | 2697.44 | ▇▁▁▁▁ |
| Ammonium, Filtered (NH4) | 319411 | 0.10 | 0.53 | 1.86 | 0.00 | 0.01 | 0.02 | 0.25 | 76.39 | ▇▁▁▁▁ |
| Nitrite (NO2) | 307591 | 0.13 | 0.03 | 0.14 | 0.00 | 0.00 | 0.00 | 0.01 | 14.10 | ▇▁▁▁▁ |
| Nitrate (NO3) | 322088 | 0.09 | 0.35 | 3.51 | -0.01 | 0.00 | 0.01 | 0.04 | 216.06 | ▇▁▁▁▁ |
| NO2+3, Filtered | 292945 | 0.17 | 0.27 | 2.63 | 0.00 | 0.00 | 0.01 | 0.06 | 218.00 | ▇▁▁▁▁ |
| Phosphate, Filtered (PO4) | 311180 | 0.12 | 0.15 | 0.28 | 0.00 | 0.00 | 0.01 | 0.10 | 2.60 | ▇▁▁▁▁ |
| pH | 290791 | 0.18 | 4.05 | 4.67 | 0.00 | 1.00 | 1.78 | 7.68 | 820.00 | ▇▁▁▁▁ |
| Specific Conductivity | 351520 | 0.01 | 2600.53 | 9322.27 | 0.00 | 45.30 | 49.56 | 54.89 | 53465.00 | ▇▁▁▁▁ |
| Turbidity | 285975 | 0.19 | 2.54 | 5.47 | 0.00 | 0.60 | 1.00 | 2.69 | 254.00 | ▇▁▁▁▁ |
| Total Kjeldahl Nitrogen | 300334 | 0.15 | 0.46 | 0.40 | 0.00 | 0.18 | 0.36 | 0.62 | 9.20 | ▇▁▁▁▁ |
| Total Phosphorus | 303465 | 0.14 | 0.03 | 0.08 | 0.00 | 0.01 | 0.01 | 0.03 | 3.38 | ▇▁▁▁▁ |
| Total Nitrogen | 322127 | 0.09 | 0.69 | 3.90 | 0.00 | 0.20 | 0.34 | 0.55 | 296.21 | ▇▁▁▁▁ |
| Chlorophyll a, Uncorrected for Pheophytin | 350110 | 0.01 | 0.62 | 2.09 | 0.04 | 0.22 | 0.32 | 0.49 | 54.60 | ▇▁▁▁▁ |
| Dissolved Oxygen Saturation | 353751 | 0.00 | 93.37 | 16.77 | 36.23 | 84.46 | 94.53 | 100.72 | 147.89 | ▁▂▇▂▁ |
| Total Suspended Solids | 349939 | 0.01 | 4.46 | 6.88 | 0.02 | 1.10 | 2.03 | 4.40 | 83.00 | ▇▁▁▁▁ |
| Nitrogen, organic | 353999 | 0.00 | 32.90 | 26.56 | 0.00 | 17.32 | 27.23 | 45.02 | 292.07 | ▇▁▁▁▁ |
| Chlorophyll a, Corrected for Pheophytin | 334548 | 0.06 | 3.70 | 9.42 | 0.00 | 0.39 | 0.97 | 3.22 | 747.00 | ▇▁▁▁▁ |
| Ammonia, Un-ionized (NH3) | 341599 | 0.04 | 0.81 | 0.30 | 0.00 | 0.55 | 1.00 | 1.00 | 1.00 | ▁▁▂▁▇ |
To make analysis easier, an unique ID is given to each row to identify the spatial location. This identifier can be built from {source}-{site}-{depth}.
df_wide$site_id <- paste(
df_wide$source,
df_wide$site,
ifelse(is.na(df_wide$sample_depth), "NA", df_wide$sample_depth),
sep = "-"
)
# Display first 100 rows with scrolling
DT::datatable(
head(df_wide, 100),
options = list(
scrollX = TRUE,
scrollY = "400px",
pageLength = 10
),
caption = "Wide format data with site_id column (scroll to see all columns)"
)# desired remaining columns: analytes, datetime, site_id, depth
# Get analyte column names (all columns except the id columns)
id_columns <- c("source", "site", "units", "latitude", "longitude", "sample_depth", "site_id", "datetime")
analyte_columns <- setdiff(names(df_wide), id_columns)
# Keep only datetime, site_id, sample_depth, and all analyte columns
df_wide <- df_wide %>%
select(site_id, datetime, sample_depth, all_of(analyte_columns))
# Display first 100 rows with scrolling
DT::datatable(
head(df_wide, 100),
options = list(
scrollX = TRUE,
scrollY = "400px",
pageLength = 10
),
caption = "Cleaned wide format data with selected columns (scroll to see all columns)"
)id_columns <- c("source", "site", "units", "latitude", "longitude", "sample_depth", "site_id", "datetime")
analyte_columns <- setdiff(names(df_wide), id_columns)
library(dplyr)
library(tidyr)
library(knitr)
df_wide %>%
summarise(across(all_of(analyte_columns), ~ sum(!is.na(.)))) %>%
pivot_longer(
cols = everything(),
names_to = "Analyte",
values_to = "Valid_Count"
) %>%
kable(caption = "Number of Valid Values per Analyte")| Analyte | Valid_Count |
|---|---|
| Water Temperature | 91069 |
| Salinity | 87863 |
| Dissolved Oxygen | 77800 |
| Ammonium, Filtered (NH4) | 34828 |
| Nitrite (NO2) | 46648 |
| Nitrate (NO3) | 32151 |
| NO2+3, Filtered | 61294 |
| Phosphate, Filtered (PO4) | 43059 |
| pH | 63448 |
| Specific Conductivity | 2719 |
| Turbidity | 68264 |
| Total Kjeldahl Nitrogen | 53905 |
| Total Phosphorus | 50774 |
| Total Nitrogen | 32112 |
| Chlorophyll a, Uncorrected for Pheophytin | 4129 |
| Dissolved Oxygen Saturation | 488 |
| Total Suspended Solids | 4300 |
| Nitrogen, organic | 240 |
| Chlorophyll a, Corrected for Pheophytin | 19691 |
| Ammonia, Un-ionized (NH3) | 12640 |
Summary of numeric analyte columns:| Name | df_wide[, numeric_cols] |
| Number of rows | 354239 |
| Number of columns | 21 |
| _______________________ | |
| Column type frequency: | |
| numeric | 21 |
| ________________________ | |
| Group variables | None |
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| sample_depth | 198581 | 0.44 | 0.44 | 0.20 | 0.00 | 0.50 | 0.50 | 0.50 | 1.00 | ▃▁▇▂▁ |
| Water Temperature | 263170 | 0.26 | 19.55 | 11.63 | 0.00 | 2.10 | 24.60 | 28.45 | 74.65 | ▃▇▂▁▁ |
| Salinity | 266376 | 0.25 | 18.52 | 16.10 | 0.00 | 1.16 | 22.72 | 34.57 | 933.00 | ▇▁▁▁▁ |
| Dissolved Oxygen | 276439 | 0.22 | 4.45 | 11.33 | 0.00 | 1.51 | 5.05 | 6.45 | 2697.44 | ▇▁▁▁▁ |
| Ammonium, Filtered (NH4) | 319411 | 0.10 | 0.53 | 1.86 | 0.00 | 0.01 | 0.02 | 0.25 | 76.39 | ▇▁▁▁▁ |
| Nitrite (NO2) | 307591 | 0.13 | 0.03 | 0.14 | 0.00 | 0.00 | 0.00 | 0.01 | 14.10 | ▇▁▁▁▁ |
| Nitrate (NO3) | 322088 | 0.09 | 0.35 | 3.51 | -0.01 | 0.00 | 0.01 | 0.04 | 216.06 | ▇▁▁▁▁ |
| NO2+3, Filtered | 292945 | 0.17 | 0.27 | 2.63 | 0.00 | 0.00 | 0.01 | 0.06 | 218.00 | ▇▁▁▁▁ |
| Phosphate, Filtered (PO4) | 311180 | 0.12 | 0.15 | 0.28 | 0.00 | 0.00 | 0.01 | 0.10 | 2.60 | ▇▁▁▁▁ |
| pH | 290791 | 0.18 | 4.05 | 4.67 | 0.00 | 1.00 | 1.78 | 7.68 | 820.00 | ▇▁▁▁▁ |
| Specific Conductivity | 351520 | 0.01 | 2600.53 | 9322.27 | 0.00 | 45.30 | 49.56 | 54.89 | 53465.00 | ▇▁▁▁▁ |
| Turbidity | 285975 | 0.19 | 2.54 | 5.47 | 0.00 | 0.60 | 1.00 | 2.69 | 254.00 | ▇▁▁▁▁ |
| Total Kjeldahl Nitrogen | 300334 | 0.15 | 0.46 | 0.40 | 0.00 | 0.18 | 0.36 | 0.62 | 9.20 | ▇▁▁▁▁ |
| Total Phosphorus | 303465 | 0.14 | 0.03 | 0.08 | 0.00 | 0.01 | 0.01 | 0.03 | 3.38 | ▇▁▁▁▁ |
| Total Nitrogen | 322127 | 0.09 | 0.69 | 3.90 | 0.00 | 0.20 | 0.34 | 0.55 | 296.21 | ▇▁▁▁▁ |
| Chlorophyll a, Uncorrected for Pheophytin | 350110 | 0.01 | 0.62 | 2.09 | 0.04 | 0.22 | 0.32 | 0.49 | 54.60 | ▇▁▁▁▁ |
| Dissolved Oxygen Saturation | 353751 | 0.00 | 93.37 | 16.77 | 36.23 | 84.46 | 94.53 | 100.72 | 147.89 | ▁▂▇▂▁ |
| Total Suspended Solids | 349939 | 0.01 | 4.46 | 6.88 | 0.02 | 1.10 | 2.03 | 4.40 | 83.00 | ▇▁▁▁▁ |
| Nitrogen, organic | 353999 | 0.00 | 32.90 | 26.56 | 0.00 | 17.32 | 27.23 | 45.02 | 292.07 | ▇▁▁▁▁ |
| Chlorophyll a, Corrected for Pheophytin | 334548 | 0.06 | 3.70 | 9.42 | 0.00 | 0.39 | 0.97 | 3.22 | 747.00 | ▇▁▁▁▁ |
| Ammonia, Un-ionized (NH3) | 341599 | 0.04 | 0.81 | 0.30 | 0.00 | 0.55 | 1.00 | 1.00 | 1.00 | ▁▁▂▁▇ |
library(dplyr)
library(tidyr)
library(ggplot2)
# 1. compute analyte order (most -> least)
analyte_order <- df_wide %>%
summarise(across(all_of(analyte_columns), ~ sum(!is.na(.)))) %>%
pivot_longer(everything(), names_to = "Analyte", values_to = "Count") %>%
arrange(desc(Count)) %>%
pull(Analyte)
# 2. presence matrix using sorted analytes
presence <- df_wide %>%
mutate(across(all_of(analyte_order),
~ !is.na(suppressWarnings(as.numeric(.))))) %>%
select(all_of(analyte_order))
# 3. co-occurrence matrix (rows/cols are in analyte_order)
cooccurrence_matrix <- t(presence) %*% as.matrix(presence)
# 4. tidy it and set factor levels explicitly
cooccurrence_df <- as.data.frame(cooccurrence_matrix) %>%
mutate(Analyte1 = row.names(.)) %>%
pivot_longer(-Analyte1,
names_to = "Analyte2",
values_to = "Cooccurrence") %>%
mutate(
Analyte1 = factor(Analyte1, levels = analyte_order),
Analyte2 = factor(Analyte2, levels = analyte_order)
) %>%
# Keep only the lower triangle (row >= column)
filter(as.numeric(Analyte1) <= as.numeric(Analyte2))
# 5. plot with y-axis reversed
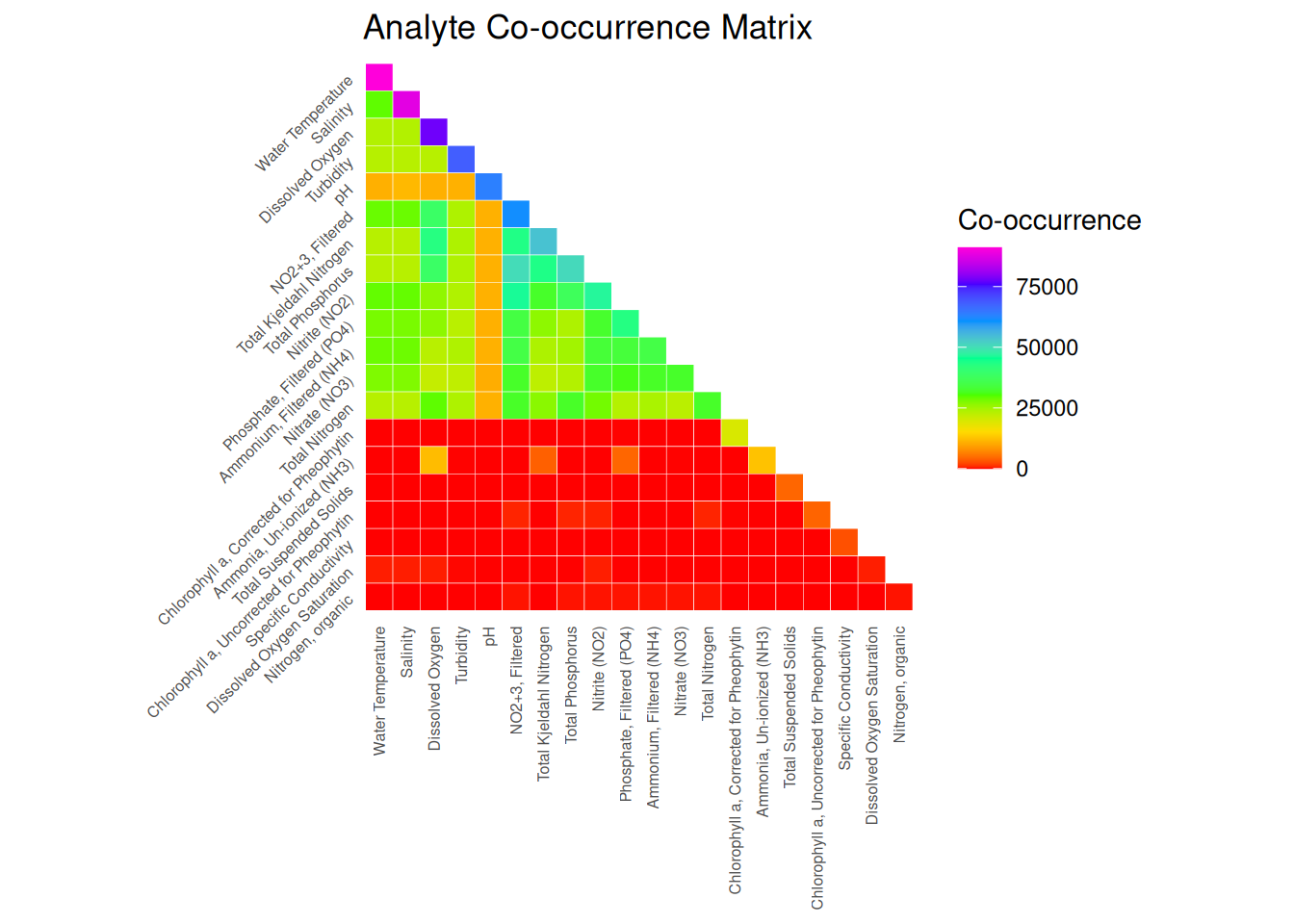
ggplot(cooccurrence_df, aes(x = Analyte1, y = Analyte2, fill = Cooccurrence)) +
geom_tile(color = "white") +
scale_fill_gradientn(colors = rainbow(7)) +
scale_y_discrete(limits = rev(levels(cooccurrence_df$Analyte2))) + # <-- reverse y-axis
labs(
title = "Analyte Co-occurrence Matrix",
x = NULL,
y = NULL,
fill = "Co-occurrence"
) +
coord_fixed() +
theme_minimal() +
theme(
axis.text.x = element_text(angle = 90, size = 6, vjust = 0.5, hjust = 1),
axis.text.y = element_text(angle = 45, size = 6, hjust = 1),
panel.grid = element_blank()
)
Note: Ideas for next steps: - Multivariate analysis where you need multiple parameters as separate variables - Creating correlation matrices between different analytes - Machine learning tasks that expect features as columns - Quick comparison of multiple parameters at the same observation point